The University of California, San Francisco (UCSF) research scientist Mansour Abtahi, PhD, was featured in the February Center for Intelligent Imaging (ci2) SRG Pillar Meeting. His presentation, "Multimodal Foundation Models for Prostate Cancer," discussed the use of artificial intelligence (AI) in the detection of prostate cancer.
Based on the location of the prostate gland and its proximity to the bladder, kidneys, spleen and small intestine, it is of high risk to spread to other organs, making early detection critical. By 2040, the global number of men with prostate cancer is expected to double. The detection of prostate cancer has revolutionized with the development of PSMA (Prostate-Specific Membrane Antigen) PET imaging, which can spot nodal and distant metastases, reduces the need for repeat biopsies and can help guide the surgery or radiation treatment.
The creation of Abtahi's foundation model included a large-scale model trained on text, medical images, audio and video data. The multimodal foundation model consists of contrastive, self-prediction, generative and generative visual language models. In addition to the LLMs, Abtahi and his team developed task-specific algorithms for data segmentation and analysis.
Between 2016-2024, Abtahi and his research team collected the metadata and radiology reports from 4,735 PSMA/PET CT pairs, 2,071 PSMA/MRI pairs and 372 PSMA/PET CT pairs with lesion annotations.
In an analysis of the foundation model's performance, they found that the model was learning the characteristics of prostate cancer.
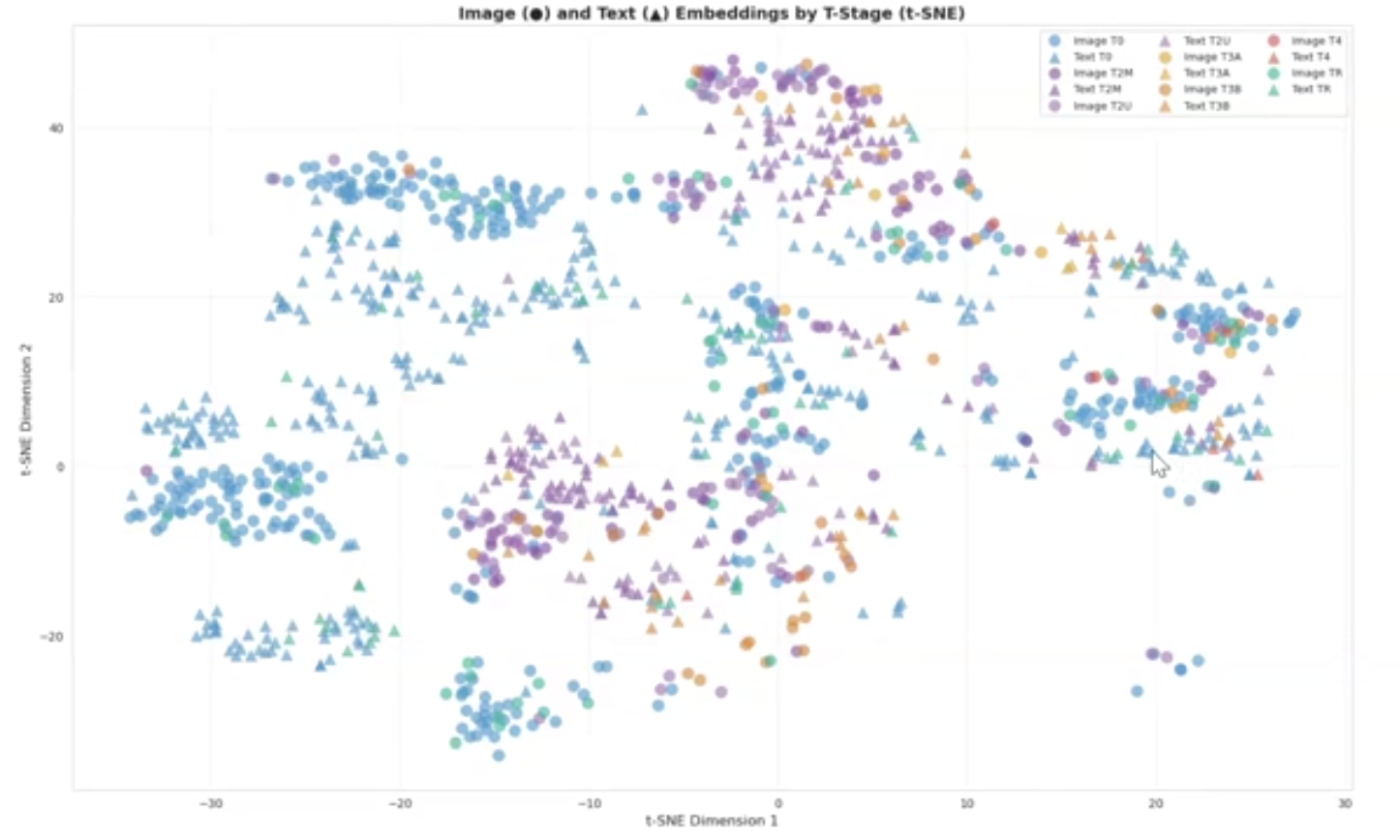
"For different stages, we have some colonies that are related to different stages, and this shows that the model has learned features that are related to different stages of prostate cancer," Abtahi says.
Through the development of the multimodal foundation model, "the model learned aligned vision and language representations and enabled good zero-shot miTNM staging," Abtahi says. "Results showed that this model has the potential to be used for different downstream (models)."
In the future, Abtahi would like to evaluate the foundation models on smaller segmentation datasets and test other functions to better optimize the cancer detection rate.
Visit the ci2 events page to learn more about the upcoming SRG Pillar meetings.



